|
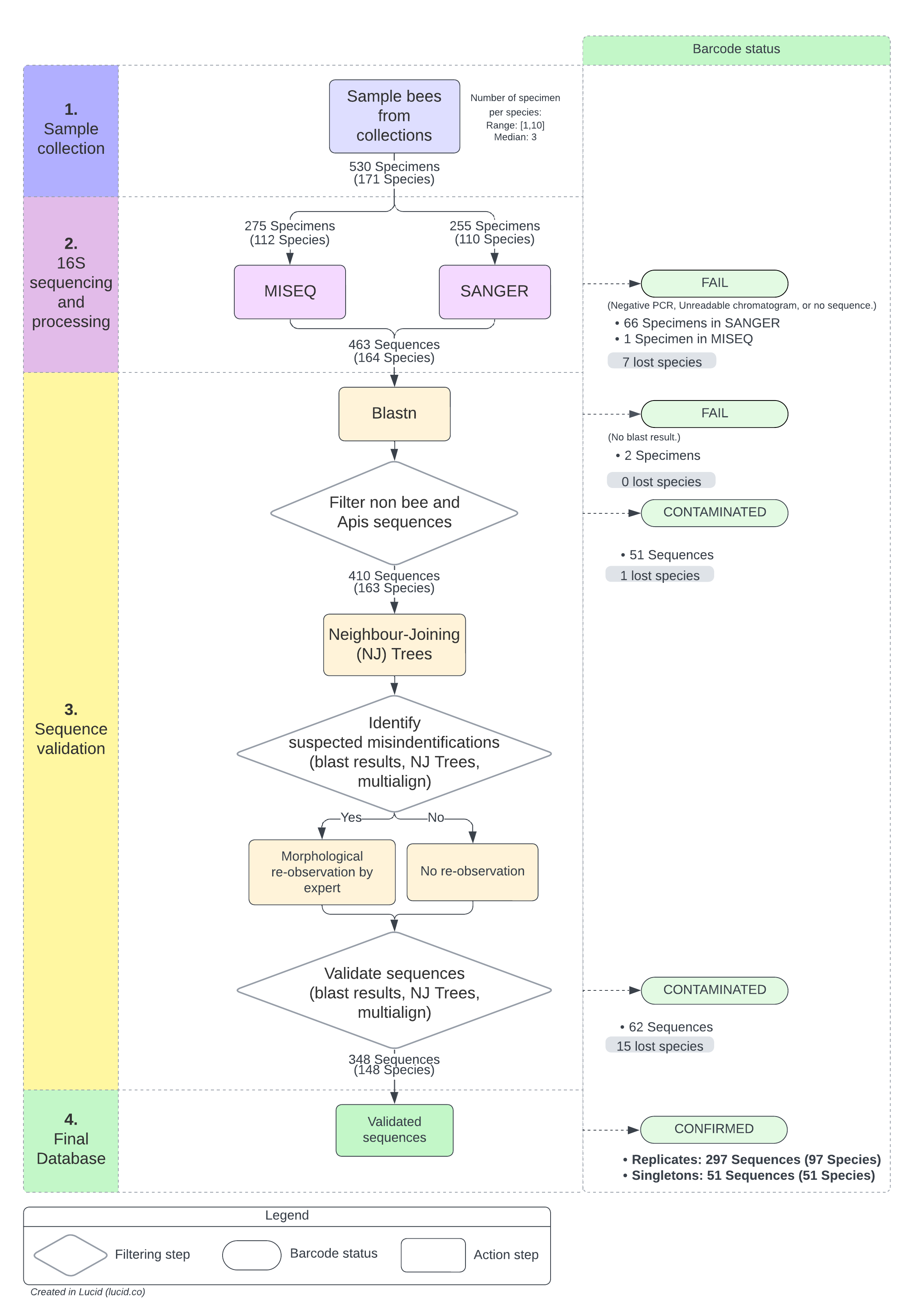
16S mini-barcode library workflow, from sampling to validation. There are four main steps represented by different colours in the left panel, from top to bottom. The middle part of the figure represent the workflow. The right panel indicates the final barcode status. For sequence validation, we used : sequence assignation by BLASTn (Altschul et al. 1990) on GenBank nt database (Sayers et al. 2021); Neighbour-joining (NJ) distance-tree inferences using the K2P model and the Muscle algorithm (Edgar 2004) for alignment implemented in the BOLD toolkit; Sequence alignment using multalin software to visualise allelic variations (Corpet 1988). For some species, the default BLAST parameters were adjusted to take into account for the high AT content of the region in this genus.
|